At one level this framework of links and objects can be put together out of existing pieces from online tools and services but on another the existing tools are totally inadequate. While their are a wide range of freely accessible sites for hosting data of different or arbitrary types, documents, and bookmarks, and these can be linked together in various ways there are very few tools and services that provide the functionality of managing links within the kind of user friendly environment that would be like to encourage adoption. Most existing web tools have also been built to be "sticky" so as to keep users on the site. This means that they are often not good at providing functionality to link out to objects on other services.
The linked data web-native notebook described above could potentially be implemented using existing tools. A full implementation would involve a variety of structured documents placed on various services using specified controlled vocabularies. The relationships between these documents would then be specified by either an XML feed generated via some other service, or by a feed generated by depositions to a triple store. Either of these approaches would then require a controlled vocabulary or vocabularies to be used in description of relationships and therefore the feed.
In practice, while this is technically feasible, for the average researcher the vocabularies are often not available or not appropriate. The tools for generating semantic documents, whether XML or RDF based are not, where they exist at all, designed with the general user in mind. The average lab is therefore restricted to a piecemeal approach based on existing, often general consumer web services. This approach can go some distance, using wikis, online documents, and data visualization extensions. An example of this approach is described in (Bradley et al., 2009) where a combination of Wikis, GoogleDoc Spreadsheets and visualization tools based on the GoogleChart API were used in a distributed set of pages that linked data representations to basic data to the procedures used to generate it through simple links. However, it currently can’t exploit the full potential of a semantic approach.
Clearly there is a gap in the tool set for delivering a linked experimental record. But what is required to fill that gap? High quality dataservices are required and are starting to appear in specific areas with a range of business models. Many of those that exist in the consumer space already provide the type of functionality that would be required including RSS feeds, visualization and management tools, tagging and categorization, and embedding capabilities. Slideshare and Flickr are excellent models for scientific data repositories in many ways.
Sophisticated collaborative online and offline document authoring tools are available. Online tools include blogs and wikis, and increasingly offline tools including Microsoft Word and Open Office, provide a rich and useable user experience that is well integrated with online publishing systems. Tools such as ICE can provide a sophisticated semantic authoring environment making it possible to effectively link structured information together.
The missing link
What is missing are the tools that will make it easy to connect items together. Semantic authoring systems solve part of the problem by enabling the creation of structured documents and in some cases by assisting in the creation of links between objects. However these are usually inward looking. The key to the web-native record is that it is integrated into the wider web, monitoring sites and feeds for new objects that may need to be incorporated into the record. These monitoring services would then present these objects to the user within a semantic authoring tool, ideally in a contextual manner.
The conceptually simplest system would monitor appropriate feeds for new objects and present these to the user as possible inputs and outputs. The user would then select an appropriate set of inputs and outputs and select the relationship between them from a limited set of possible relationships (is an input to, is an output of, generated data). This could be implemented as three drop down menus in it's simplest form but this would only apply after the event. Such a tool would not be particularly useful in planning experiments or as a first pass recording tool and would therefore add another step to the recording process.
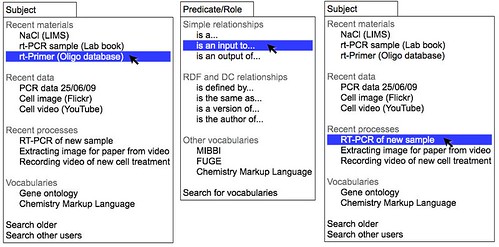
Figure 2. A conceptual tool for connecting web objects together via simple relationships. A range of web services are monitored to identify new objects, data, processes, documents, or controlled vocabulary terms that may be relevant to the uesr. In this simple tool these are presented as potential subjects and objects in drop down down menus. The relationship can be be selected from a central menu. The output of the tool is a feed of relationships between web accessible objects.
Capturing in silico process: A new type of log file
In an ideal world, the whole process of creating the linked record would be done behind the scenes, without requiring the intervention of the user. This is likely to be challenging in the experimental laboratory but is entirely feasible for in silico work and data analysis within defined tools. Many data analysis tools already generate log files, although as data analysis tools have become more GUI driven these have become less obvious to the user and more focussed on aiding in technical support. Within a given data analysis or computational tool objects will be created and operated on by procedures hard coded into the system. The relationships are therefore absolute and defined.
The extensive work of the reproducible research movement on developing approaches and standards to recording and communicating computational procedures has largely focussed on the production of log files and command records (or scripts) that can be used to reproduce an analysis procedure as well as arguing for the necessity to provide running code and the input and intermediate data files. In the linked record world it will be necessary to create one more "logfile" that describes the relationships between all the objects created by reference to some agreed vocabulary. This "relationships logfile" which would ideally be RDF or a similar framework is implicit in a traditional log file or script but by making it explicit it will be possible to wire these computational processes into a wider web of data automatically. Thus the area of computational analysis is where the most rapid gains can be expected to be made as well as where the highest standards are likely to be possible. The challenge of creating similar log files for experimental research is greater, but will benefit significantly from building up experience in the in silico world.
Systems for capturing physical processes
For physical experimentation it is possible to imagine an authoring tool that automatically tracks feeds of possible input and output objects as the researcher describes what they are planning or what they are doing. The authoring tool would trigger a plugin or internal system to identify points where new links should be made, based on the document that the researcher is generating as they plan or execute their experiment. For example, typing the sentence, “an image was taken of the sample for treatment A” would trigger the system to look at recent items from the feed (or feeds) of the appropriate image service(s), which in turn would be presented to the user in a drop down menu for selection. The selection of the correct item would add a link from the document to the image. The “sample for treatment A” having already been defined a statement would then be incorporated in a machine readable form within the document that “sample for treatment A” was the input for the process “an image was taken” which generated data (the image).
Such a system would consist of two parts; first an intelligent feed reader which monitors all the relevant feeds, laboratory management systems for samples, data repositories, or laboratory instruments, for new data. In a world in which it seems natural that the Mars Phoenix lander should have a Twitter account, and indeed the high throughput sequencers at the Sanger Centre send status updates via Twitter, the notion of an instrument automatically providing a status update on the production of data seems natural. What are less prevalent are similar feeds generated by sample and laboratory information management systems although the principles are precisely equivalent; when an object (data or sample or material) is created or added to the system a feed item is created and pushed out for notification.
The second part of the system is more challenging. Providing a means of linking inputs to outputs, via for example drop down menus is relatively straightforward. Even the natural language processing required to automatically recognise where links should be created is feasible with todays technology. But capturing what those links mean is more difficult. There is a balance that needs to be found between providing sufficient flexibility to describe a very wide range of possible connections ("is an input to", "is the data generated from") and providing enough information, via a rigorously designed controlled vocabulary, to enable detailed automated parsing by machines. Where existing vocabularies and ontologies exist and are directly applicable to the problem in hand these should clearly be used and strongly supported by the authoring tools. In the many situations where they are not ideal then more flexibility needs to be supported allowing the researcher to go "off piste" but at the same time ambiguity should be avoided. Thus such a system needs to "nudge" the user into using the most appropriate available structured description of their experiment, while allowing them the choice to use none, but at the same time helping them avoid using terms or descriptions that could be misintepreted. Not a trivial system to build.









